Division of Cancer Cell Regulation
Introduction
We seek to understand the mechanisms that maintain cellular homeostasis and their dysfunction in cancer. Normal cellular homeostasis requires the coordinated regulation of signaling molecules in space, time, and quantity. Accumulation of genetic and epigenetic alterations or aberrant extracellular stimuli disrupt the stringent regulation of signaling networks and lead to cellular transformation and tumor progression. Our studies involve dissecting genes, proteins, and signaling mechanisms directly responsible for oncogenic phenotypes and identifying novel therapeutic targets. Currently, the goals of our research are to determine the molecular mechanisms underlying aberrant activation of the Src pathway in a wide variety of human cancer cells.
Research topics
We seek to understand the mechanisms maintaining cellular homeostasis and their dysfunction in cancer. Normal cellular homeostasis requires the coordinated regulation of signaling molecules in space, time, and quantity. Accumulation of genetic and epigenetic alterations or oncogenic viral infections disrupts the stringent regulation of signaling networks and leads to cellular transformation and tumor progression. Our studies involve dissecting the genes, proteins, and signaling mechanisms directly responsible for oncogenic phenotypes and identifying novel therapeutic targets. Currently, the goals of our research are to elucidate the molecular mechanisms underlying aberrant activation of Src pathways in a wide variety of human cancer cells.
Elucidating the molecular mechanisms of exosome biogenesis
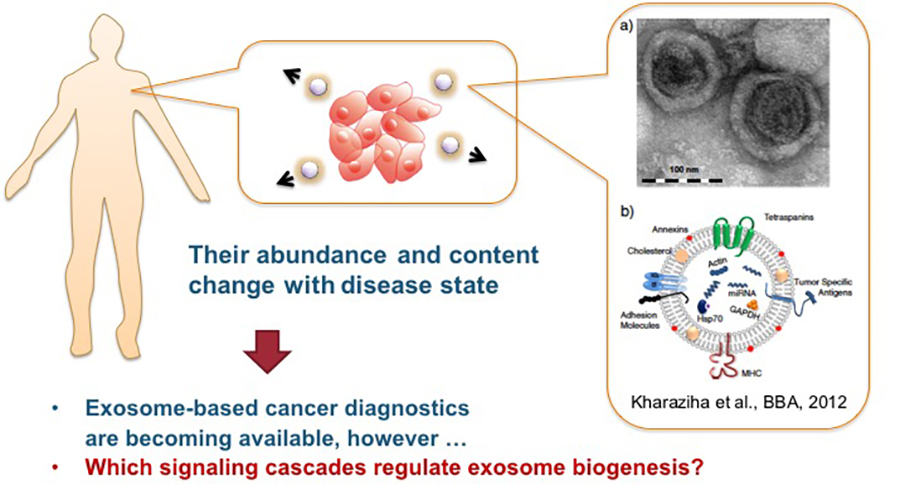
Exosomes are extracellular secretory vesicles that are thought to play important roles in intercellular communication. The amount and composition of exosomes vary depending on the physiological state of the cells such as cancers, but the mechanism remains unclear (See Figure 1). We found that exosome secretion is enhanced by the kinase activity of c-Src. c-Src is a non-receptor tyrosine kinase, which is frequently overexpressed or hyper-activated in various human cancers. To elucidate the molecular mechanisms underlying c-Src-mediated exosome secretion, we explored c-Src-interacting proteins in exosomes by mass-spectrometry. Among the several candidates, Alix, an ESCRT-associated protein, interacted with c-Src via SH3 domain-mediated binding in exosome. Inhibition of Src activity with dasatinib treatment suppressed exosome secretion and shRNA-mediated Alix knockdown. These data indicate that Alix regulates c-Src-mediated exosome production. Further, we are currently developing a new analytical method for exosomes and clarifying the mechanisms underlying the regulation of exosome biogenesis by identifying and analyzing molecules, such as Alix, responsible for exosome formation and encapsulation (Hikita et al., Sci Rep, 2018).
One of the representative studies published can be accessed at the linked below:
Sensitive and rapid quantification of exosomes by fusing luciferase to exosome marker proteins
The project is supported by JST, PRESTO, a funding agency in Japan.
Elucidating the spatial regulation of oncogenic signaling via lipid rafts
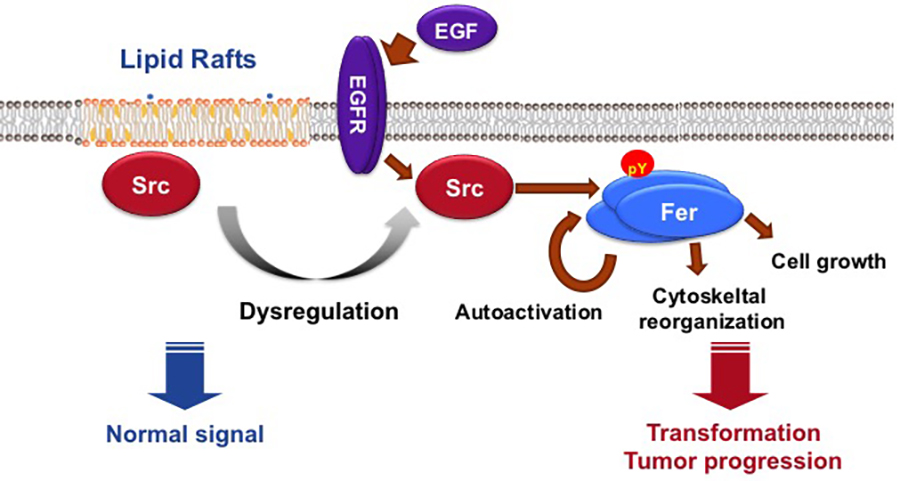
c-Src is upregulated in various human cancers, suggesting that it plays a role in malignant progression. However, the molecular circuits of c-Src oncogenic signaling remain unclear. We demonstrated that Fer tyrosine kinase oligomer mediates and amplifies Src-induced tumor progression. Previously, we showed that transformation of fibroblasts is promoted by relocation of c-Src to non-raft membranes (Oneyama et al., Mol Cell, 2008; MCB, 2009). Under these conditions, we identified Fer and ezrin as non-raft c-Src targets (Oneyama et al., Oncogene, 2016). c-Src directly activated Fer by initiating its autophosphorylation, which was further amplified by Fer oligomerization. Fer interacted with active c-Src at focal adhesion membranes and activated Fer-phosphorylated ezrin to induce cell transformation. Fer was also crucial for cell transformation induced by v-Src or epidermal growth-factor receptor activation. Furthermore, Fer activation was required for tumorigenesis and invasiveness in some cancer cells in which c-Src is upregulated. We propose that the Src?Fer axis represents a new therapeutic target for treating a subset of human cancers (See Figure 2).
One of the representative studies published can be accessed at the link below:
Fer tyrosine kinase oligomer mediates and amplifies Src-induced tumor progression
The project is supported by AMED, a funding agency in Japan.
Clarifying the roles of micro(mi)RNAs in cancer progression
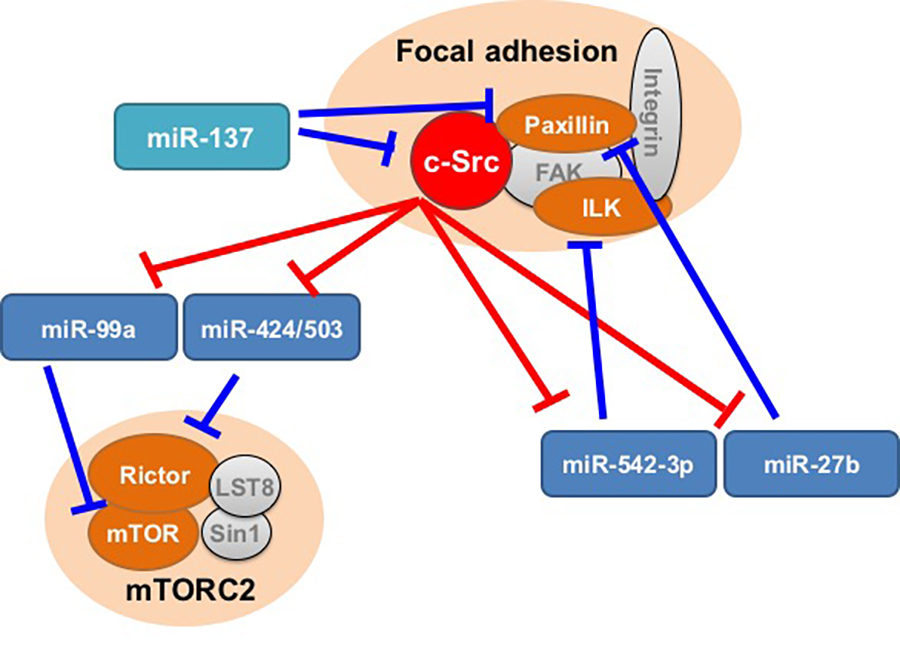
The tyrosine kinase c-Src is frequently overexpressed and activated in a wide variety of human cancers. However, the molecular mechanisms underlying c-Src-mediated tumor progression remain unclear. To verify the function of c-Src in cancer progression, we previously developed an experimental system using Csk-deficient mouse embryonic fibroblasts in which c-Src induces cellular transformation (Oneyama et al., Genes Cells). Using this system, the expression profiles of miRNAs between non-transformed and c-Src-transformed cells were compared. Microarray profiling revealed that c-Src activation regulates a limited set of miRNAs; seven miRNAs were downregulated and six were upregulated by more than 2-fold (Oneyama et al., Oncogene, 2011). Studies of the function of each downregulated miRNA revealed miRNA-mediated c-Src oncogenic signaling and crosstalk between Src and other oncogenic signaling pathways, such as the mTOR and focal adhesion-mediated pathways (See Figure 3). Our studies provided insight into multiple miRNAs that work behind the scenes to regulate Src signaling. Signaling molecules, such as Src and mTOR, are linked with each other, not only via phosphorylation and inter-protein interactions but also via translational regulation by miRNAs. This interplay may lead to novel insights into cancer progression, invasiveness, and drug resistance.
Our published review can be accessed at the link below:
MicroRNAs as the fine-tuners of Src oncogenic signalling
Members
1995 M.S., Graduate School of Science, Osaka University
1995-2003 Research associate, Tokyo Research Laboratories, Kyowa Hakko Kogyo Co., Ltd.
2003 PhD., Graduate School of Medicine, Osaka University
2003-2005 Postdoctoral Fellow, Osaka Bioscience Institute
2005-2007 Postdoctoral Fellow, Research Institute for Microbial Diseases, Osaka University
2007-2011 Assistant Professor, Research Institute for Microbial Diseases, Osaka University
2011-2015 Associate Professor, Research Institute for Microbial Diseases, Osaka University
2015-present Chief, Aichi Cancer Center Research Institute
2016-present Visiting Professor, Graduate School of Pharmaceutical Sciences, Nagoya City University
2017-2021 Researcher, JST PRESTO “Fine Particles”
2018-present Visiting Professor, Graduate School of Medicine, Nagoya University
2020-present Visiting Professor, Institute of Nano-Life-Systems, Nagoya University
Analysis of stress responses and secretion inhibition signaling using extracellular vesicles as markers
mechanisms of risk factors, expanding the possibilities for new molecular targets and drug selection.
2009 M.S. Graduate School of Pharmaceutical Sciences, Osaka University
2012 PhD., Graduate School of Pharmaceutical Sciences, Osaka University
2012-2015 Specially Appointed Postdoctoral Fellow, WPI Immunology Frontier Research Center (IFReC), Osaka University
2015-2016 Specially Appointed Postdoctoral Fellow, Graduate School of Medical Sciences, Kanazawa University
2016-2018 Specially Appointed Assistant Professor, Graduate School of Medical Sciences, Kanazawa University
2018-2024 Specially Appointed Assistant Professor, WPI Nano Life Science Institute (NanoLSI), Kanazawa University
2021-2024 Specially Appointed Assistant Professor, WISE Program for Nano-Precision Medicine, Science, and Technology, Kanazawa University
2024-2025 Team Leader, Chemical Manufacturer
Through research that builds a bridge from the movements of tiny molecules to life-saving medical care, I want to contribute to society.
Publications
Original article
- Mitani F, Hayasaka R, Hirayama A, *Oneyama C. SNAP23-Mediated Perturbation of Cholesterol-Enriched Membrane Microdomain Promotes Extracellular Vesicle Production in Src-Activated Cancer Cells. Biol Pharm Bull. 45(10):1572-1580, 2022. (PMID: 36184518)
- Hikita T, *Oneyama C. Quantification and Imaging of Exosomes via Luciferase-Fused Exosome Marker Proteins: ExoLuc System. Methods Mol Biol. 2524:281-290, 2022. (PMID: 35821479)
- Mitani F, Lin J, Sakamoto T, Uehara R, Hikita T, Yoshida T, Setiawan A, Arai M, *Oneyama C. Asteltoxin inhibits extracellular vesicle production through AMPK/mTOR-mediated activation of lysosome function. Sci Rep. 12(1):6674, 2022. (PMID: 35461323)
- Hikita T, Uehara R, Itoh RE, Mitani F, Miyata M, Yoshida T, Yamaguchi R, *Oneyama C. MEK/ERK-mediated oncogenic signals promote secretion of extracellular vesicles by controlling lysosome function. Cancer Sci. 113(4):1264-1276, 2022. (PMID: 35108425)
- Morioka S, Nakanishi H, Yamamoto T, Hasegawa J, Tokuda E, Hikita T, Sakihara T, Kugii Y, Oneyama C, Yamazaki M, Suzuki A, Sasaki J, Sasaki T. A mass spectrometric method for in-depth profiling of phosphoinositide regioisomers and their disease-associated regulation. Nat Commun. 13(1):83, 2022. (PMID: 35013169)
- Kunou S, Shimada K, Takai M, Sakamoto A, Aoki T, Hikita T, Kagaya Y, Iwamoto E, Sanada M, Shimada S, Hayakawa F, Oneyama C, Kiyoi H. Exosomes secreted from cancer-associated fibroblasts elicit anti-pyrimidine drug resistance through modulation of its transporter in malignant lymphoma. Oncogene, 40(23):3989-4003, 2021. (PMID: 33994542)
- Ito RE, Oneyama C, Aoki K. Oncogenic mutation or overexpression of oncogenic KRAS or BRAF is not sufficient to confer oncogene addiction. PLoS One, 16(4): e0249388, 2021. (PMID: 33793658)
- Nishimura T, Oyama T, Hu HT, Fujioka T, Hanawa-Suetsugu K, Ikeda K, Yamada S, Kawana H, Saigusa D, Ikeda H, Kurata R, Oono-Yakura K, Kitamata M, Kida K, Hikita T, Mizutani K, Yasuhara K, Mimori-Kiyosue Y, Oneyama C, Kurimoto K, Hosokawa Y, Aoki J, Takai Y, Arita M, Suetsugu S. Filopodium-derived vesicles produced by MIM enhance the migration of recipient cells. Dev Cell, 56(6): 842-859.e8, 2021. (PMID: 33756122)
- Hikita T, Miyata M, Watanabe R, *Oneyama C. In vivo imaging of long-term accumulation of cancer-derived exosomes using a BRET-based reporter. Sci Rep, 10(1): 16616, 2020. (PMID: 33024173)
- Watanabe R, Miyata M, *Oneyama C. Rictor promotes tumor progression of rapamycin-insensitive triple-negative breast cancer cells. Biochem Biophys Res Commun, 531(4):636-642, 2020. (PMID: 32819718)
- Okuzaki D, Yamauchi T, Mitani F, Miyata M, Ninomiya Y, Akamatsu H, *Oneyama C. c-Src promotes tumor progression via downregulation of miR-129-1-3p. Cancer Science, 111(2): 418-428, 2020 (PMID: 31799727)
- Hikita T, Kuwahara A, Watanabe R, Miyata M, *Oneyama C.: Src in endosomal membranes promotes exosome secretion and tumor progression. Sci Rep, 9(1): 3265, 2019. (PMID: 30824759)
- Hikita T, Miyata M, Watanabe R, *Oneyama C.: Sensitive and rapid quantification of exosomes by fusing luciferase to exosome marker proteins. Sci Rep, 8(1): 14035, 2018. (PMID: 30232365)
- Kokuda R, Watanabe R, Okuzaki D, Akamatsu H, *Oneyama C.: MicroRNA-137-mediated Src oncogenic signaling promotes cancer progression. Genes Cells, 23(8): 688-701, 2018. (PMID: 29962093)
- *Oneyama C, Yoshikawa Y, Ninomiya Y, Iino T, Tsukita S, Okada M.: Fer tyrosine kinase oligomer mediates and amplifies Src-induced tumor progression. Oncogene, 35(4): 501-512, 2016. (PMID: 25867068)
- Matsuyama R, Okuzaki D, Okada M, *Oneyama C.: miR-27b suppresses tumor progression by regulating ARFGEF1 and the focal adhesion signaling. Cancer Science, 107(1): 28-35, 2016. (PMID: 26473412)
- Kakumoto K, Ikeda J, Okada M, Morii E, *Oneyama C.: mLST8 promotes mTOR-mediated tumor progression. PLoS One, 10(4): e0119015, 2015. (PMID: 25906254)
- Kajiwara K, Yamada T, Bamba T, Fukusaki E, Imamoto F, Okada M, *Oneyama C.: c-Src-induced activation of ceramide metabolism impairs membrane microdomains and promotes malignant progression by facilitating the translocation of c-Src to focal adhesions. Biochem J, 458(1): 81-93, 2014. (PMID: 24266736)
- *Oneyama C, Kito Y, Asai R, Ikeda J, Yoshida T, Okuzaki D, Kokuda R, Kakumoto K, Takayama K, Inoue S, Morii E, Okada M.: MiR-424/503-mediated Rictor upregulation promotes tumor progression. PLoS One, 8(11): e80300, 2013. (PMID: 24244675)
- *Oneyama C, Morii E, Okuzaki D, Takahashi Y, Ikeda J, Wakabayashi N, Akamatsu H, Tsujimoto M, Nishida T, Aozasa K, Okada M.: MicroRNA-mediated upregulation of integrin-linked kinase is crucial for Src-induced tumor progression. Oncogene, 31(13), 1623-1635, 2012. (PMID: 21860426)
- *Oneyama C, Ikeda J, Okuzaki D, Suzuki K, Kanou T, Shintani Y, Morii E, Okumura M, Aozasa K, Okada M.: MicroRNA-mediated downregulation of mTOR/FGFR3 controls tumor growth induced by Src-related oncogenic pathways. Oncogene, 30(32): 3489-3501, 2011. (PMID: 21383697)
- Kuroiwa M, Oneyama C, Nada S, Okada M.: The guanine nucleotide exchange factor Arhgef5 plays crucial roles in c-Src-induced podosome formation. J Cell Sci, 124: 1726-1738, 2011. (PMID: 21525037)
- Suzuki K, Oneyama C, Kimura H, Tajima S, Okada M.: Downregulation of the tumor suppressor Cbp/PAG1 is mediated by epigenetic histone modifications via the MAPK/PI3K pathway. J Biol Chem, 286(18): 15698-15706, 2011. (PMID: 21388951)
- Kanou T, Oneyama C, Kawahara K, Okimura A, Ohta M, Ikeda N, Shintani Y, Okumura M, Okada M.: The transmembrane adaptor Cbp/PAG1 controls the malignant potential of human non-small cell lung cancers that have c-src upregulation. Mol Cancer Res, 9(1): 103-114, 2011. (PMID: 21156787)
- Hikita T, Oneyama C, Okada M.: Purvalanol A, a CDK inhibitor, effectively suppresses Src-mediated transformation by inhibiting both CDKs and c-Src. Genes Cells, 15(10): 1051-1062, 2010. (PMID: 20825494)
- Oneyama C, Iino T, Saito K, Suzuki K, Ogawa A, Okada M.: Transforming potential of Src family kinases is limited by the cholesterol-enriched membrane microdomain. Mol Cell Biol, 29(24): 6462-6472, 2009. (PMID: 19822664)
- Inoue K, Sone T, Oneyama C, Nishiumi F, Kishine H, Sasaki Y, Andoh T, Okada M, Chesnut JD, Imamoto F.: A versatile nonviral vector system for tetracycline-dependent one-step conditional induction of transgene expression. Gene Therapy, 16(12): 1383-1394, 2009. (PMID: 19759563)
- Oneyama C, Hikita T, Enya K, Dobenecker MW, Saito K, Nada S, Tarakhovsky A, Okada M. : The lipid raft-anchored adaptor protein Cbp controls the oncogenic potential of c-Src. Mol Cell, 30(4): 426-436, 2008. (PMID: 18498747)
- Saito K, Enya K, Oneyama C, Hikita T, Okada M.: Proteomic identification of ZO-1/2 as a novel scaffold for Src/Csk regulatory circuit. Biochem Biophys Res Commun, 366(4): 969-975, 2008. (PMID: 18086565)
- Oneyama C, Hikita T, Nada S, Okada M.: Functional dissection of transformation by c-Src and v-Src. Genes Cells, 13(1): 1-12, 2008. (PMID: 18173743)
- Yagi R, Waguri S, Sumikawa Y, Nada S, Oneyama C, Itami S, Schmedt C, Uchiyama Y, Okada M.: C-terminal Src kinase controls development and maintenance of mouse squamous epithelia. EMBO J, 26(5): 1234-1244, 2007. (PMID: 17304209)
- Sukezane T, Oneyama C, Kakumoto K, Shibutani K, Hanafusa H and Akagi T.: Human diploid fibroblasts are resistant to MEK/ERK-mediated disruption of the actin cytoskeleton and invasiveness stimulated by Ras. Oncogene, 24: 5648-5655, 2005. (PMID: 16007212)
- Oneyama C, Agatsuma T, Kanda Y, Nakano H, Sharma SV, Nakano H, Narazaki F and Tatsuta K.: Synthetic Inhibitors of Proline-Rich Ligand-Mediated Protein-Protein Interaction: Potent Analogs of UCS15A. Chemistry & Biology, 10: 443-451, 2003. (PMID: 12770826)
- Oneyama C, Nakano H and Sharma SV.: UCS15A, a novel small molecule, SH3 domain-mediated protein-protein interaction blocking drug. Oncogene, 21: 2037-2050, 2002. (PMID: 11960376)
- Sharma SV, Oneyama C, Yamashita Y, Nakano H, Sugawara K, Hamada M, Kosaka N and Tamaoki T.: UCS15A, a non-kinase inhibitor of Src signal transduction. Oncogene, 20: 2068-2079, 2001. (PMID: 11360191)
Education & Training
Our laboratory is a part of the Nagoya University Graduate School of Medicine and Nagoya City University Graduate School of Pharmaceutical Sciences. Foreign graduate students are welcome to study in our programs, including those who are supported by the Japanese Government Monbusho (MEXT) scholarship. For details regarding entrance examinations and scholarships, please follow this link.

