Division of Molecular Therapeutics
Introduction
We have been focusing on developing effective therapies against cancers with aberrant MPAK activation, especially caused by RAS/RAF mutations. We also aim to expand the molecular classification of colorectal cancer by integrating ctDNA analysis results, whole exome and RNA sequencing data.
Research topics
Resistance Mechanism to KRAS inhibitors
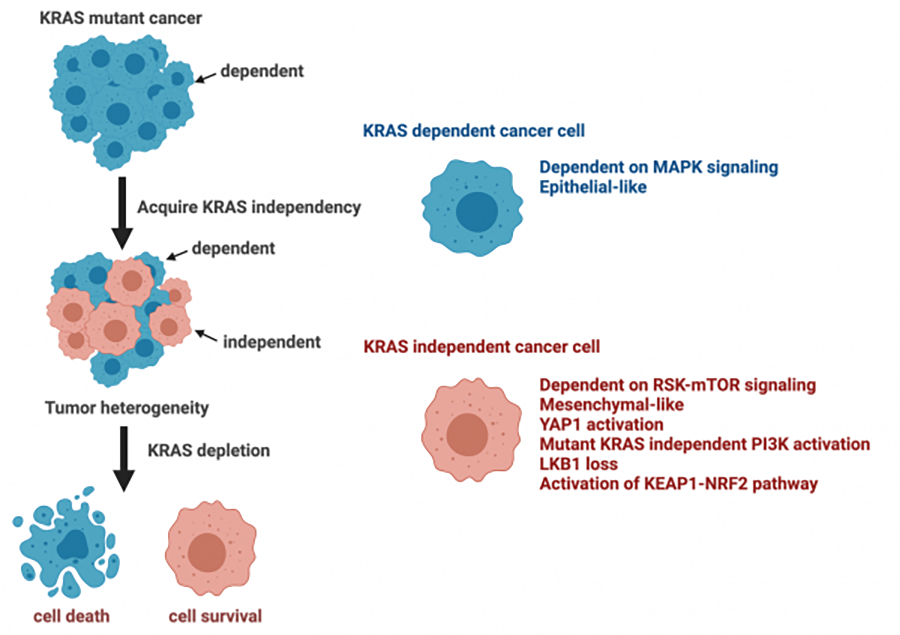
KRAS mutations are commonly observed in cancers. The relatively smooth surface architecture of RAS and its picomolar affinity for GTP/GDP have given rise to the assumption that RAS is “undruggable”. However, recent identification of a pocket in its switch II region has allowed targeting of tumors with the KRAS G12C mutation. While the observed responses in KRAS-mutant patients, especially with lung cancer, are encouraging, the response rate is low compared with other drugs targeting driver oncogenes such as EGFR and ALK inhibitors. We have elucidated that tumor cells can acquire mutant KRAS independence by activating pathways that functionally substitute for the mutant KRAS. These KRAS-independent tumor cells exhibit a mesenchymal phenotype, readily primed for potential metastasis. Activation of YAP and mutations in LKB1, KEAP1, and/or NRF2, associated with mutant KRAS autonomy, rewire survival signaling and metabolic processes. The presence of KRAS-independent cells is associated with the heterogeneity of KRAS mutant cancers, as well as variable responses to therapies. Notably, KRAS G12C-specific inhibitors appear to be effective only in tumors dependent on mutant KRAS for their survival. Therefore, determining KRAS dependence will be critical for selecting patients who should be treated with mutant-specific inhibitors. Furthermore, elucidating underlying mechanisms of KRAS autonomy is crucial for developing optimal treatment strategies for KRAS-independent tumors.
Mutli-omics analysis for colorectal cancer: integrating the results of circulating tumor DNA, exome and transcriptome analyses
The current standard of care for patients with colorectal cancer includes surgical resection followed by adjuvant chemotherapy. All stage III and pathologically assessed high-risk stage II patients are treated with 5-FU based chemotherapy. However, more than half are estimated to be free from risk of recurrence even without adjuvant chemotherapy. Therefore, identification of predictive markers for recurrence is necessary to avoid unnecessary treatment and reduce medical costs. In addition, molecular targets for patients demonstrating relapse despite receiving adjuvant chemotherapy are a high priority. The detection of circulating tumor DNA in blood is a relatively non-invasive method that may help genotyping, selection of treatment options, and monitoring response to treatment. To investigate the potential use of ctDNA markers for detecting residual disease to predict who is at high risk of relapse, and using ctDNA assay as a guide for adjuvant chemotherapy, a multicenter investigator-initiated trial, CIRCULATE-Japan, was launched in May 2020 and more than 4000 patients who have undergone curative surgery for colorectal cancer have been prospectively enrolled. Our laboratory has responsibility for translational research within this project, integrating ctDNA, mutational, and RNA sequencing analyses to identify molecular subtypes and new molecular targets. Our twin goal is to establish biomarkers to stratify patients who do not require adjuvant chemotherapy and identify molecular targets to enhance the efficacy of adjuvant chemotherapy.
Members
2006 Ph.D. from Nagoya City University
2007-2008 Postdoctoral Fellow at division of carcinogenesis in Nagoya University
2008-2012 Research Fellow at Jeffrey Engelman Laboratory at Massachusetts General Hospital Cancer Center
2012-2016 Assistant Professor, Division of Medical Oncology, Cancer Research Institute, Kanazawa University.
2016-2018 Associate Professor, Division of Medical Oncology, Cancer Research Institute, Kanazawa University
2018-present Chief, Division of Molecular Therapeutics, Aichi Cancer Center Research Institute
2018/4-2018/10 Visiting Scientist, Novartis Institute for Biomedical Research (Cambridge MA, USA).
2007-2011 Postdoctoral Fellow, Duke University
2011-2015 Designated Assistant Professor, Graduate School of Medicine, Center for Neural Disease and Cancer Division, Nagoya University
2015-2018 Assistant Professor, Graduate School of Medicine, Center for Research of Laboratory Animals and Medical Research Engineering Division for Advanced Medical Research, Nagoya University
2018-Present. Senior Scientist, Division of Molecular Diagnostics, Aichi Cancer Center
Publications
2024
- Kimura R, Adachi Y, Hirade K, Kisoda S, Yanase S, Shibata N, Ishii M, Fujiwara Y, Yamaguchi R, Fujita Y, Hosoda W, Ebi H. ARAF Amplification in Small-Cell Lung Cancer-Transformed Tumors Following Resistance to Epidermal Growth Factor Receptor-Tyrosine Kinase Inhibitors. Cancers. 16:3501, 2024.
- Bando H, Yamaguchi K, Mitani S, Sawada K, Mishima S, Komine K, Okugawa Y, Hosoda W, Ebi H. Japanese Society of Medical Oncology clinical guidelines: Molecular testing for colorectal cancer treatment, 5th edition. Cancer Sci. 115:1014-1021, 2024.
- Kitai H, Ebi H. Oncogene alterations in non-small cell lung cancer with FGFR1 amplification-novel approach to stratify patients who benefit from FGFR inhibitors. Transl Lung Cancer Res. 13:684-688, 2024.
- Kitai H, Choi P, Yang Y, Boyer J, Whaley A, Pancholi P, Thant C, Reiter J, Chen K, Markov V, Dr Taniguchi H, Yamaguchi R, Ebi H, Evans J, Jiang J, Lee B, Wildes D, de Stanchina E, Smith J, Dr Singh , Rosen N. Combined inhibition of KRASG12C and mTORC1 kinase in non-small cell lung cancer causes synergistic cell death and tumor regression. Nature Communications. 15:6076, 2024.
- Iida N, Imai M, Okamoto W, Kato T, Esaki T, Kato K, Komatsu Y, Yuki S, Masuishi T, Nishina T, Ebi H (11/27). Novel ERBB2 Variant Potentially Associated with Resistance against Anti-HER2 Monoclonal Antibody-Based Therapy in ERBB2-Amplified Metastatic Colorectal Cancer. Clin Cancer Res. 30:4167-4178, 2024
- Nakamura Y, Watanabe J, Akazawa N, Hirata K, Kataoka K, Yokota M, Kato K, Kotaka M, Kagawa Y, Yeh KH, Mishima S, Yukami H, Ando K, Miyo M, Misumi T, Yamazaki K, Ebi H (17/36). ctDNA-based molecular residual disease and survival in resectable colorectal cancer. Nat Med. 30:3272-3283, 2024.
- Nakano Y, Masuda T, Sakamoto T, Tanaka N, Tobo T, Hashimoto M, Tatsumi T, Saito H, Takahashi J, Koike K, Abe T, Ando Y, Ozato Y, Hosoda K, Hirose K, Higuchi S, Ikehara T, Hisamatsu Y, Toshima T, Yonemura Y, Ogino T, Uemura M, Eguchi H, Doki Y, Mimori K. SHARPIN is a novel gene of colorectal cancer that promotes tumor growth potentially via inhibition of p53 expression. Int J Oncol. 65(6):113, 2024.
- Rubinson DA*, Tanaka N*, Fece de la Cruz F, Kapner KS, Rosenthal MH, Norden BL, Barnes H, Ehnstrom S, Morales-Giron AA, Brais LK, Lemke CT, Aguirre AJ, Corcoran RB. Sotorasib Is a Pan-RASG12C Inhibitor Capable of Driving Clinical Response in NRASG12C Cancers. Cancer Discov. 14(5):727-736, 2024 (* equal contribution)
2023
- Adachi Y, Kimura R, Hirade K, Yanase S, Nishioka Y, Kasuga N, Yamaguchi R, Ebi H. Scribble mis-localization induces adaptive resistance to KRAS G12C inhibitors through feedback activation of MAPK signaling mediated by YAP-induced MRAS. Nature Cancer. 2023 4:829-843, 2023.
- Ebi H. Drug-Tolerant Persister Cells After EGFR Tyrosine Kinase Inhibitor Treatment: Their Origin and the Influences From the Tumor Microenvironment. Journal of Thoracic Oncology. 18:399-401, 2023.
- Maeda A, Ando H, Irie K, Hashimoto N, Morishige JI, Fukushima S, Ebi H, Uchida K, Iwata H, Sawaki M. Effects of ABCB1 and ABCG2 Polymorphisms on the Pharmacokinetics of Abemaciclib Metabolites (M2, M20, M18). Anticancer Res. 43:1283-1289, 2023.
- Hiramatsu K, Matsuda C, Masago K, Toriyama K, Sasaki E, Fujita Y, Haneda M, Ebi H, Shibata N, Hosoda W. Diagnostic utility of DNA integrity number as an indicator of sufficient DNA quality in next-generation sequencing-based genomic profiling. Am J Clin Pathol. 11:aqad 046, 2023
- Yoshino T, Cervantes A, Bando H, Martinelli E, Oki E, Ebi H. (27/33) Pentheroudakis G. Pan-Asian adapted ESMO Clinical Practice Guidelines for the diagnosis, treatment and follow-up of patients with metastatic colorectal cancer. ESMO Open. 8:101588, 2023.
- Matsubara J, Mukai K, Kondo T, Yoshioka M, Kage H, Oda K, Kudo R, Ikeda S, Ebi H, Muro K, Hayashi R, Tokudome N, Yamamoto N, Muto M*. First-Line Genomic Profiling in Previously Untreated Advanced Solid Tumors for Identification of Targeted Therapy Opportunities. JAMA Netw Open. 2023;6:e2323336
- Lai X, Lui SKL, Lam HY, Adachi Y, Sim WJ, Vasilevski N, Armstrong NJ, Bridgeman SC, Main NM, Tan TZ, Tirnitz-Parker JEE, Thiery JP, Ebi H*, Kumar AP*, Eichhorn PJA*. SHP2 inhibitors maintain TGFβ signalling through SMURF2 inhibition. NPJ Precis Oncol. 7:136, 2023. (*Co-corresponding author)
- Iwai M*, Kajino T*, Nakatochi M, Yanagisawa K, Hosono Y, Isomura H, Shimada Y, Suzuki M, Taguchi A, Takahashi T. Long non-coding RNA TILR constitutively represses TP53 and apoptosis in lung cancer. Oncogene. 42(5):364-373, 2023. (*: equal contribution)
- Tanaka N*, Okada H*, Yamaguchi K, Seki M, Matsubara D, Gotoh N, Suzuki Y, Furukawa Y, Yamashita T, Inoue JI, Kaneko S, Sakamoto T. Mint3-depletion-induced energy stress sensitizes triple-negative breast cancer to chemotherapy via HSF1 inactivation. Cell Death Dis. 14(12):815, 2023
- Tanaka N, Sakamoto T. MT1-MMP as a Key Regulator of Metastasis. Cells. 12(17):2187, 2023
- Tanaka N, Sakamoto T. Mint3 as a Potential Target for Cooling Down HIF-1α-Mediated Inflammation and Cancer Aggressiveness. Biomedicines. 11(2):549, 2023
2022
- Cai J, Jacob S, Kurupi R, Dalton KM, Coon C, Greninger P, Egan RK, Stein GT, Murchie E, McClanaghan J, Adachi Y, Hirade K, Dozmorov M, Glod J, Boikos SA, Ebi H, Hao H, Caponigro G, Benes CH, Faber AC. High-risk neuroblastoma with NF1 loss of function is targetable using SHP2 inhibition. Cell Reports, 40:111095, 2022
- Su, WJ; Mukherjee, R; Yaeger, R; Son, J; Xu, JN; Na, N; Timaul, NM; Hechtman, J; Paroder, V; Lin, M; Mattar, M; Qiu, J; Chang, Q; Zhao, HY; Zhang, J; Little, M; Adachi, Y; Han, SW; Taylor, BS; Ebi, H; Abdel-Wahab, O; de Stanchina, E; Rudin, CM; Janne, PA; McCormick, F; Yao, Z; Rosen, N. ARAF protein kinase activates RAS by antagonizing its binding to RASGAP NF1. Molecular Cell, 82:2443-2457, 2022.
- Vega P.N., Nilsson A., Kumar M.P., Niitsu H., Simmons A.J., Ro J., Wang J., Chen Z., Joughin B.A., Li W., McKinley E.T., Liu Q., Roland J.T., Washington M.K., Coffey R.J., Lauffenburger D.A., and Lau K.S. Cancer-Associated Fibroblasts and Squamous Epithelial Cells Constitute a Unique Microenvironment in a Mouse Model of Inflammation-Induced Colon Cancer. Front Oncol,12;878920, 2022.
2021
- Escaping KRAS: Gaining Autonomy and Resistance to KRAS Inhibition in KRAS Mutant Cancers. Cancers. 13:5081, 2021.
- Nakamura, Y; Okamoto, W; Kato, T; Esaki, T; Kato, K; Komatsu, Y; Yuki, S; Masuishi, T; Nishina, T; Ebi, H; Sawada, K; Taniguchi, H; Fuse, N; Nomura, S; Fukui, M; Matsuda, S; Sakamoto, Y; Uchigata, H; Kitajima, K; Kuramoto, N; Asakawa, T; Olsen, S; Odegaard, JI; Sato, A; Fujii, S; Ohtsu, A; Yoshino, T. Circulating tumor DNA-guided treatment with pertuzumab plus trastuzumab for HER2-amplified metastatic colorectal cancer: a phase 2 trial. Nature Med. 27:1899-1903, 2021.
- Nakasuka, F; Tabata, S; Sakamoto, T; Hirayama, A; Ebi, H; Yamada, T; Umetsu, K; Ohishi, M; Ueno, A; Goto, H; Sugimoto, M; Nishioka, Y; Yamada, Y; Tomita, M; Sasaki, AT; Yano, S; Soga, T. TGF-beta-dependent reprogramming of amino acid metabolism induces epithelial-mesenchymal transition in non-small cell lung cancers. Communications Biology, 4:782, 2021.
- Taniguchi, H; Nakamura, Y; Kotani, D; Yukami, H; Mishima, S; Sawada, K; Shirasu, H; Ebi, H; Yamanaka, T; Aleshin, A; Bllings, PR; Rabinowitz, M; Oki, E; Takemasa, I; Kato, T; Mori, M; Yoshino, T. CIRCULATE-Japan: Circulating tumor DNA-guided adaptive platform trials to refine adjuvant therapy for colorectal cancer. Cancer Science, 112:2915-2920, 2021.
- Niitsu H., Lu Y., Huh W.J., Love A.M., Franklin J.L., and Coffey R.J., Cell-Autonomous Role of EGFR in Spontaneous Duodenal Tumors in LRIG1 Null Mice. Cell Mol Gastroenterol Hepatol, 12: 1159-1162, 2021.
- Chen B., Scurrah C.R., McKinley E.T., Simmons A.J., Ramirez-Solano M.A., Zhu X., Markham N.O., Heiser C.N., Vega P.N., Rolong A., Kim H., Sheng Q., Drewes J.L., Zhou Y., Southard-Smith A.N., Xu Y., Ro J., Jones A.L., Revetta F., Berry L.D., Niitsu H., Islam M., Pelka K., Hofree M., Chen J.H., Sarkizova S., Ng K., Giannakis M., Boland G.M., Aguirre A.J., Anderson A.C., Rozenblatt-Rosen O., Regev A., Hacohen N., Kawasaki K., Sato T., Goettel J.A., Grady W.M., Zheng W., Washington M.K., Cai Q., Sears C.L., Goldenring J.R., Franklin J.L., Su T., Huh W.J., Vandekar S., Roland J.T., Liu Q., Coffey R.J., Shrubsole M.J., and Lau K.S., Differential pre-malignant programs and microenvironment chart distinct paths to malignancy in human colorectal polyps. Cell, 184:6262-6280, 2021.
2020
- Adachi, Y; Ito, K; Hayashi, Y; Kimura, R; Tan, TZ; Yamaguchi, R; Ebi, H. Epithelial-to-Mesenchymal Transition is a Cause of Both Intrinsic and Acquired Resistance to KRAS G12C Inhibitor in KRAS G12C-Mutant Non-Small Cell Lung Cancer. Clinical Cancer Res, 26:5962-5973, 2020.
- Costa C, Wang Y, Ly A, Hosono Y, Ellen M, Walmsley CS, Huynh T, Healy C, Peterson R, Yanase S, Jakubik CT, Henderson LE, Damon LJ, Timonina D, Sanidas I, Pinto CJ, Mino-Kenudson M, Stone JR, Dyson NJ, Ellisen LW, Bardia A, Ebi H, Benes CH, Engelman JA and Juric D. PTEN loss mediates clinical cross-resistance to CDK4/6 and PI3Kalpha inhibitors in breast cancer. Cancer Discov. 10, 72-85, 2020.
- Ebi H, Bando H, Taniguchi H, Sunakawa Y, Okugawa Y, Hatanaka Y, Hosoda W, Kumamoto K, Nakatani K, Yamazaki K. Japanese Society of Medical Oncology Clinical Guidelines: Molecular Testing for Colorectal Cancer Treatment, 4th edition. Cancer Sci. 111:3962-3969, 2020
- Huh W.J., Niitsu H., Carney B., McKinley E.T., Houghton J.L., and Coffey R.J., Identification and Characterization of Unique Neutralizing Antibodies to Mouse EGF Receptor. Gastroenterology, 158: 1500-1502, 2020.
2019 selected
- Yaeger R, Kotani D, Mondaca S, Parikh AR, Bando H, Van Seventer EE, Taniguchi H, Zhao H, Thant CN, de Stanchina E, Rosen N, Corcoran RB, Yoshino T, Yao Z and Ebi H. Response to Anti-EGFR Therapy in Patients with BRAF non-V600-Mutant Metastatic Colorectal Cancer. Clin Cancer Res. 25:7089-7097, 2019.
2018 selected
- Song KA, Hosono Y, Turner C, Jacob S, Lochmann TL, Murakami Y, Patel NU, Ham J, Hu B, Powell KM, Coon CM, Windle BE, Oya Y, Koblinski JE, Harada H, Leverson JD, Souers AJ, Hata AN, Boikos S, Yatabe Y, Ebi H* and Faber AC*. Increased Synthesis of MCL-1 Protein Underlies Initial Survival of EGFR-Mutant Lung Cancer to EGFR Inhibitors and Provides a Novel Drug Target. Clin Cancer Res. 24:5658-5672, 2018. (*Co-corresponding author)
- Kotani H, Adachi Y, Kitai H, Tomida S, Bando H, Faber AC, Yoshino T, Voon DC, Yano S and Ebi H. Distinct dependencies on receptor tyrosine kinases in the regulation of MAPK signaling between BRAF V600E and non-V600E mutant lung cancers. Oncogene. 37:1775-1787, 2018.
Education & Training
Positions for Ph.D. students and Postdoctoral Fellows
Postdoctoral positions are available for highly motivated candidates with a background in molecular and cellular biology to join our lab. The focus of our laboratory research program is to investigate the molecular mechanisms of dysregulated cellular signaling in pathways playing critical roles in tumor cell survival. For this purpose, we inhibit activity of molecules of interest using small molecule inhibitors, knock out/down or overexpression. Results of our research are being translated into clinical settings.

