Division of Molecular Genetics
Introduction
We are conducting genome-wide genomic and epigenomic analyses of human cancers using state-of-the-art technologies, including next-generation sequencing (NGS), microarray, and bioinformatics, as well as identifying novel cancer-related genes as possible diagnostic biomarkers/therapeutic targets. In our group, research interests range from germline genetic variants related to susceptibility for cancers to somatic alterations associated with the development/progression/malignant phenotypes of cancers, such as epithelial-to-mesenchymal transition (EMT), metastasis, and drug resistance. Because tumor heterogeneity is related to tumor progression, metastasis, recurrence, and drug resistance, quantitative detection of genetic and epigenetic alterations in tissues as well as liquid biopsy samples and their application in evaluation of tumor heterogeneity are being assessed. These approaches will contribute to development of (1) medicines for preventing cancers based on personalized risk assessment and intervention, (2) personalized therapies for cancers by overcoming tumor heterogeneity based on genomic and epigenomic information, and (3) novel non-invasive/less-invasive diagnostic methods for detecting diagnostic/therapeutic effect determination/prognostic markers for cancers.
Research topics
Identification and functional annotation of novel cancer-related genes/variants by detecting cancer-specific genomic/epigenomic/transcriptomic alterations as landmarks
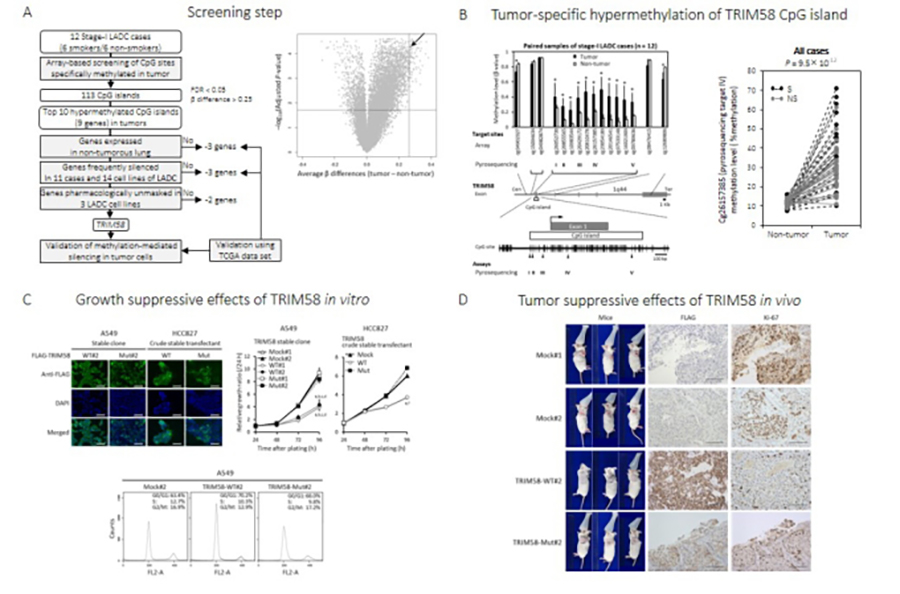
Genome-wide genomic, epigenomic, and transcriptomic analyses of human cancers using state-of-the-art technologies, including next-generation sequencing (NGS), microarray, and bioinformatics, can identify novel cancer-related genes, which may be useful as diagnostic biomarkers/therapeutic targets in human cancers. Clinicopathological and biological factors can be assessed using clinical samples and cell/animal-based functional assays, respectively. Recently, we identified TRIM58 as a novel tumor-suppressor gene for lung adenocarcinoma (LADC), which is methylated and downregulated in the early phase of LADC and suppresses cancer cell growth upon forced expression in vitro and in vivo (Figure 1; Kajiura et al., 2017). A similar approach can be used with publicly available databases, such as TCGA and GEO (Kohmoto et al., unpublished data). To identify useful and highly reliable diagnostic biomarkers/therapeutic targets at a clinical site, we are developing rapid and efficient bioinformatics/experimental pipelines using various methods including genome editing (Mitsui et al., 2016) for functional annotation of candidate genes and variants. We have identified more than 50 cancer-related genes in various types of cancers, such as esophageal and oral squamous cell carcinoma, gastric cancer, non-small cell lung cancer, and gynecological cancers, using this approach.
Development of quantitative and/or highly sensitive methods for detecting diagnostic/therapeutic targets according to tumor heterogeneity
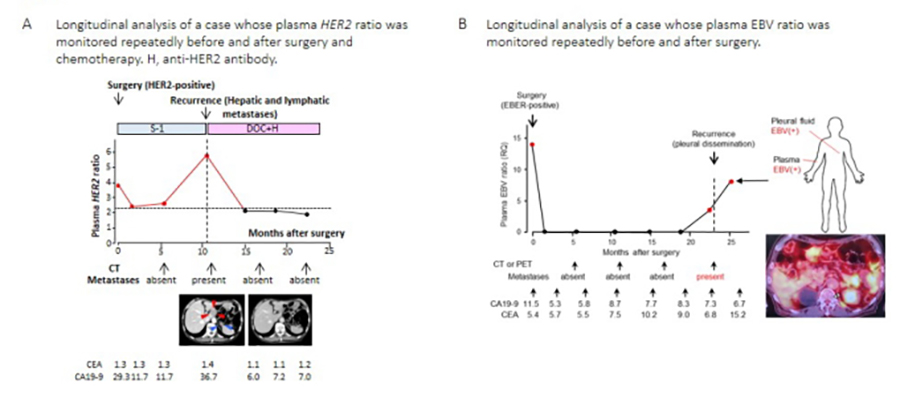
We have focused on detecting and evaluating temporal and special tumor heterogeneity and their clinical significance through quantitative and/or highly sensitive genomic/epigenomic analyses in human cancers using digital droplet PCR (ddPCR), mass spectrometry, and NGS. We use tumor biopsy or operatively extracted specimens as well as non-invasively and repetitively obtained plasma cell-free DNA (cfDNA) for this purpose. For example, we developed ddPCR-based detection methods for detecting HER2 amplification in cfDNA and demonstrated the clinical importance of monitoring HER2 amplification for therapeutic effect determination/prediction and forecasting relapse (Shoda et al., 2017a and 2017b) in gastric cancer (GC). We also determined the potential utility of Epstein?Barr virus (EBV) DNA in cfDNA as a biomarker for detecting and/or monitoring the therapeutic response in patients with EBV-associated gastric carcinoma (Figure 2; Shoda et al., 2017). In colon cancers treated with preoperative chemoradiation therapy, we demonstrated that KRAS or BRAF mutations in biopsied samples disappeared after preoperative therapy, suggesting the possibility of clonal selection by therapeutic intervention (Fujita et al., unpublished data) and usefulness of real-time detection of mutations in specific genes for selecting treatment options.
Determination of prevalence of pathogenic germline variants and development of their detection method in hereditary cancer syndrome-related genes
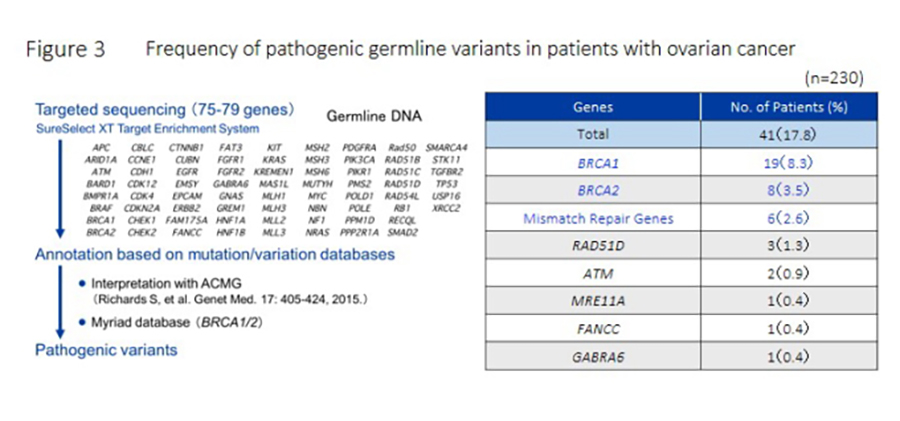
We developed detection methods and annotation pipelines for germline variants in hereditary cancer syndrome-related genes. Using these methods, we determined the prevalence of pathogenic germline variants of hereditary cancer syndrome-related genes in cases without/with controls. Recently, we determined the prevalence of pathogenic variants in primary ovarian, fallopian tube, and peritoneal carcinoma (OC) using targeted panel sequencing, identified a similar variant prevalence in Japanese patients with OC and other ethnic groups, and demonstrated that high-grade serous carcinoma and OC family history may facilitate genetic predisposition prediction in Japanese patients with OC and referring high-risk patients for genetic counseling and testing (Figure 3; Hirasawa et al., 2017). We are also collaborating with the Division of Molecular and Clinical Epidemiology and Division of Epidemiology and Prevention, ACCRI to develop methods for risk assessment, intervention, and determination of their effects on cancer prevention using genetic and environmental risk factors.
Members
Publications
Corresponding author
2020
- Osumi K, Suga K, Ono A, Goji A, Mori T, Kinoshita Y, Sugano M, Toda Y, Urushihara M, Nakagawa R, Hayabuchi Y, Imoto I, Shoji Kagami S. Molecular diagnosis of an infant with TSC2/PKD1 contiguous gene syndrome. Hum Genome Var. (in press).
- Tsuchiya M, Yamada T, Akaishi R, Hamanoue H, Hirasawa A, Hyodo M, Imoto I, Kosho T, Kurosawa K, Murakami H, Nakatani K, Nomura F, Sasaki A, Shimizu K, Tamai M, Umemura H, Watanabe A, Yoshida A, Yoshihashi H, Yotsumoto J, Kosugi S. Attitudes toward and current status of disclosure of secondary findings from next-generation sequencing: A nation-wide survey of clinical genetics professionals in Japan. J Hum Genet. 2020 Jul 13. doi: 10.1038/s10038-020-0802-2.( Online ahead of print.)
- Lin Y, Nakatochi M, Hosono Y, Ito H, Kamatani Y, Inoko A, Sakamoto H, Kinoshita F, Kobayashi Y, Ishii H, Ozaka M, Sasaki T, Matsuyama M, Sasahira N, Morimoto M, Kobayashi S, Fukushima T, Ueno M, Ohkawa S, Egawa N, Kuruma S, Mori M, Nakao H, Adachi Y, Okuda M, Osaki T, Kamiya S, Wang C, Hara K, Shimizu Y, Miyamoto T, Hayashi Y, Ebi H, Kohmoto T, Imoto I, Kasugai Y, Murakami Y, Akiyama M, Ishigaki K, Matsuda K, Hirata M, Shimada K, Okusaka T, Kawaguchi T, Takahashi M, Watanabe Y, Kuriki K, Kadota A, Okada R, Mikami H, Takezaki T, Suzuki S, Yamaji T, Iwasaki M, Sawada N, Goto A, Kinoshita K, Fuse N, Katsuoka F, Shimizu A, Nishizuka SS, Tanno K, Suzuki K, Okada Y, Horikoshi M, Yamauchi T, Kadowaki T, Yu H, Zhong J, Amundadottir LT, Doki Y, Ishii H, Eguchi H, Bogumil D, Haiman CA, Le Marchand L, Mori M, Risch H, Setiawan VW, Tsugane S, Wakai K, Yoshida T, Matsuda F, Kubo M, Kikuchi S, Matsuo K. Genome-wide association meta-analysis identifies GP2 gene risk variants for pancreatic cancer. Nat Commun. 2020 Jun 24;11(1):3175.
- Ishigaki K, Akiyama M, Kanai M, Takahashi A, Kawakami E, Sugishita H, Sakaue S, Matoba N, Low SK, Okada Y, Terao C, Amariuta T, Gazal S, Kochi Y, Horikoshi M, Suzuki K, Ito K, Koyama S, Ozaki K, Niida S, Sakata Y, Sakata Y, Kohno T, Shiraishi K, Momozawa Y, Hirata M, Matsuda K, Ikeda M, Iwata N, Ikegawa S, Kou I, Tanaka T, Nakagawa H, Suzuki A, Hirota T, Tamari M, Chayama K, Miki D, Mori M, Nagayama S, Daigo Y, Miki Y, Katagiri T, Ogawa O, Obara W, Ito H, Yoshida T, Imoto I, Takahashi T, Tanikawa C, Suzuki T, Sinozaki N, Minami S, Yamaguchi H, Asai S, Takahashi Y, Yamaji K, Takahashi K, Fujioka T, Takata R, Yanai H, Masumoto A, Koretsune Y, Kutsumi H, Higashiyama M, Murayama S, Minegishi N, Suzuki K, Tanno K, Shimizu A, Yamaji T, Iwasaki M, Sawada N, Uemura H, Tanaka K, Naito M, Sasaki M, Wakai K, Tsugane S, Yamamoto M, Yamamoto K, Murakami Y, Nakamura Y, Raychaudhuri S, Inazawa J, Yamauchi T, Kadowaki T, Kubo M, Kamatani Y. Large-scale genome-wide association study in a Japanese population identifies novel susceptibility loci across different diseases. Nat Genet. 2020;52(7):669-679.
- Sato Y, Tajima A, Kiguchi M, Kogusuri S, Fujii A, Sato T, Nozawa S, Yoshiike M, Naka-Mieno M, Kojo K, Uchida M, Tsuchiya H, Yamasaki K, Imoto I, Yamauchi A, Iwamoto T. Genome-wide association study of semen volume, sperm concentration, testis size, and plasma inhibin B levels. J Hum Genet 2020;65(8):683-691.
- Ichihara A, Yasue A, Mitsui SN, Arai D, Minegishi Y, Oyadomari S, Imoto I, Tanaka E. The C-terminal region including the MH6 domain of Msx1 regulates skeletal development. Biochem Biophys Res Commun. 2020;526(1):62-69.
- Koyanagi Y, Suzuki E, Imoto I, Kasugai Y, Isao Oze I, Ugai T, Iwase M, Usui Y, Kawakatsu Y, Sawabe M, Hirayama Y, Tanaka T, Abe T, Ito S, Komori K, Hanai N, Tajika M, Shimizu Y, Niwa Y, Ito H, Matsuo K. Across-site differences in the mechanism of alcohol-induced digestive tract carcinogenesis: an evaluation by mediation analysis. Cancer Res 2020;80(7):1601-1610.
- Kohmoto T, Masuda K, Shoda K, Takahashi R, Ujiro S, Tange S, Ichikawa D, Otsuji E, Imoto I*. Claudin-6 is a single prognostic marker and functions as a tumor-promoting gene in a subgroup of intestinal type gastric cancer. Gastric Cancer. 2020; 23(3): 403-417.
- Akahane T, Hirasawa A, Imoto I, Okubo A, Itoh M, Nanki Y, Yoshihama T, Tominaga E, Aoki D. Establishment and characterization of a new malignant peritoneal mesothelioma cell line, KOG-1, from the ascitic fluid of a patient with pemetrexed chemotherapy resistance. Hum Cell 2020;33(1):272-282.
2019
- Miyake K, Sakane A, Tsuchiya Y, Sagawa I, Tomida Y, Kasahara J, Imoto I, Watanabe S, Higo D, Mizuguchi K, Sasaki T. Actin cytoskeletal reorganization function of JRAB/MICAL-L2 is fine-tuned by intramolecular interaction between first LIM zinc finger and C-terminal coiled-coil domains. Sci Rep. 2019;9:12794.
- Mori T, Goji A,, Toda Y, Ito H, Mori K, Kohmoto T, Imoto I, Kagami S. A 16q22.2-q23.1 deletion identified in a male infant with West syndrome. Brain Dev. 2019;41(10):888-893.
- Taguchi I, Yamada T, Akaishi R, Imoto I, Kurosawa K, Nakatani K, Nomura F, Hamanoue H, Hyodo M, Murakami H, Yoshihashi H, Yotsumoto J, Kosugi S. Attitudes of clinical geneticists and certified genetic counselors to genome editing and its clinical applications: A nation-wide questionnaire survey in Japan. J Hum Genet. 2019;64(9):945-954
- Tsuboi M, Kondo K, Masuda K, Tange S, Kajiura K, Kohmoto T, Watanabe M, Takizawa H, Imoto I, Tangoku A. Prognostic significance of GAD1 overexpression in patients with resected lung adenocarcinoma. Cancer Medicine. (in press)
- Kikuchi-Koike R, Nagasaka K, Tsuda H, Ishii Y, Sakamoto M, Kikuchi Y, Fukui S, Miyagawa Y, Hiraike H, Kobayashi T, Kinoshita T, Kanai Y, Shibata T, Imoto I, Inazawa J, Matsubara O, Ayabe T. Array comparative genomic hybridization analysis discloses chromosome copy number alterations as indicators of patient outcome in lymph node-negative breast cancer. BMC Cancer. 2019;19(1):521.
- Fukuda D, Nishimoto S, Aini K, Tanaka A, Nishiguchi T, Kim-Kaneyama J, Lei XF, Masuda K, Naruto T, Tanaka K, Higashikuni Y, Hirata Y, Yagi S, Kusunose K, Yamada H, Soeki T, Imoto I, Akasaka T, Shimabukuro M, Sata M. Toll-like receptor 9 plays a pivotal role in angiotensin ll-induced atherosclerosis. J Am Heart Assoc. 2019;8(7):e010860
- Okano S, Makita Y, Katada A, Harabuchi Y, Kohmoto T, Naruto T, Masuda K, Imoto I. Novel compound heterozygous CDH23 variants in a patient with Usher syndrome type l. Hum Genome Var. 2019;6:8.
2018
- Tanikawa C, Kamatani Y, Toyoshima O, Sakamoto H, Ito H, Takahashi A, Momozawa Y, Hirata M, Fuse N, Takai-Igarashi T, Shimizu A, Sasaki M, Yamaji T, Sawada N, Iwasaki M, Tsugane S, Naito M, Hishida A, Wakai K, Furusyo N, Murakami Y, Nakamura Y, Imoto I, Inazawa J, Oze I, Sato N, Tanioka F, Sugimura H, Hirose H, Yoshida T, Matsuo K, Kubo M, Matsuda K. A GWAS identifies gastric cancer susceptibility loci at 12q24.11-1 12 and 20q11.21. Cancer Sci. 2018;109(12):4015-4024.
- Miki A, Sakurada Y, Tanaka K, Semba K, Mitamura Y, Yuzawa M, Tajima A, Nakatochi M, Yamamoto K, Matsuo K, Imoto I, Honda S. Genome-wide association study to identify a new susceptibility locus for central serous chorioretinopathy in the Japanese population. Invest Ophthalmol Vis Sci. 2018;59(13):5542-5547
- Hara T, Phuong PT, Fukuda D, Yamaguchi K, Murata C, Nishimoto S, Yagi S, Kusunose K, Yamada H, Soeki T, Wakatsuki T, Imoto I, Shimabukuro M, Sata M. Protease-activated receptor-2 plays a critical role in vascular inflammation and atherosclerosis in apolipoprotein e-deficient mice. Circulation. 2018;38(16):1706-1719.. doi: 10.1161/CIRCULATIONAHA.118.033544.
- Okamoto N, Kohmoto T, Naruto T, Masuda K, Imoto I*. Primary microcephaly caused by novel compound heterozygous mutations in ASPM. Hum Genome Var. 2018;5:18015 doi: 10.1038/hgv.2018.15
- Sato Y, Tajima A, Sato T, Nozawa S, Yoshiike M, Imoto I, Yamauchi A, Iwamoto T. Genome-wide association study identifies ERBB4 on 2q34 as a novel locus associated with sperm motility in Japanese men. J Med Genet. 2018;55(6):415-21. doi: 10.1136/jmedgenet-2017-104991.
- Enya T, Okamoto N, Iba Y, Miyazawa T, Okada M, Ida S, Naruto T, Imoto I, Fujita A, Miyake N, Matsumoto N, Sugimoto K, Takemura T. Three patients with Schaaf-Yang syndrome exhibiting arthrogryposis and endocrinological abnormalities. Am J Med Genet A. 2018;176(3):707-11. doi: 10.1002/ajmg.a.38606
- Tokaji N, Ito H, Kohmoto T, Naruto T, Takahashi R, Goji A, Mori M, Toda Y, Saito M, Tange S, Masuda K, Kagami S, Imoto I*. A rare male patient with classic Rett syndrome caused by MeCP2_e1 mutation. Am J Med Genet A. 2018;176(3):699-702. doi: 10.1002/ajmg.a.38595
- Harada R, Kimura M, Sato Y, Taniguchi T, Tomonari T, Tanaka T, Tanaka H, Muguruma N, Shinomiya H, Honda H, Imoto I, Sogabe M, Okahisa T, Takayama T. APOB codon 4311 polymorphism is associated with hepatitis C virus infection through altered lipid metabolism. BMC Gastroenterol. 2018;18:24. doi: 10.1186/s12876-018-0747-5
- Eguchi M, Ozaki E, Yamauchi T, Ohta M, Higaki T, Masuda K, Imoto I, Ishii E, Eguchi-Ishimae M. Manifestation of recessive combined D-2-, L-2-hydroxyglutaric aciduria in combination with 22q11.2 deletion syndrome. Am J Med Genet A. 2018;176:351-8. doi: 10.1002/ajmg.a.38578.
2017
- Hirasawa A, Imoto I*, Naruto T, Akahane T, Yamagami W, Nomura H, Masuda K, Susumu N, Tsuda H, Aoki D. Prevalence of pathogenic germline variants detected by multigene sequencing in unselected Japanese patients with ovarian cancer. Oncotarget. 2017;8:112258-67. doi:10.18632/oncotarget.22733
- Fujita Y, Masuda K, Hamada J, Shoda K, Naruto T, Hamada S, Miyakami Y, Kohmoto T, Watanabe M, Takahashi R, Tange S, Saito M, Kudo Y, Fujiwara H, Ichikawa D, Tangoku A, Otsuji E, Imoto I*. KH-type splicing regulatory protein is involved in esophageal squamous cell carcinoma progression. Oncotarget. 2017;8:101130-45. doi: 10.18632/oncotarget.20926.
- Kajiura K, Takizawa H, Morimoto Y, Masuda K, Tsuboi M, Kishibuchi R, Wusiman N, Sawada T, Kawakita N, Toba H, Yoshida M, Kawakami Y, Naruto T, Imoto I, Tangoku A, Kondo K. Frequent silencing of RASSF1A by DNA methylation in thymic neuroendocrine tumours. Lung Cancer. 2017;111:116-23. doi: 10.1016/j.lungcan.2017.05.019.
- Okamoto N, Kohmoto T, Naruto T, Masuda K, Komori T, Imoto I*. Novel CLCN7 compound heterozygous mutations in intermediate autosomal recessive osteopetrosis. Hum Genome Var. 2017;4:17036. doi: 10.1038/hgv.2017.36.
- Okada A, Kohmoto T, Naruto T, Yokota I, Kotani M, Shimada A, Miyamoto Y, Takahashi R, Goji A, Masuda K, Kagami S, Imoto I*. The first Japanese patient with mandibular hypoplasia, deafness, progeroid features and lipodystrophy (MDPL) diagnosed via POLD1 mutation detection. Hum Genome Var. 2017;4:17031;doi10.1038/hgv/2017.31.
- Kohmoto T, Masuda K, Naruto T, Tange S, Shoda K, Hamada J, Saito M, Ichikawa D, Tajima A, Otsuji E, Imoto I*. Construction of a combinatorial pipeline using two somatic variant calling methods for whole exome sequence data of gastric cancer. J Med Invest. 2017;64:223-240. doi: 10.2152/jmi.64.233
- Obata F, Tani K, Yamaguchi H, Tabata R, Bando H, Imoto I. Comparison of the efficacy and safety of 10-mg empagliflozin every day versus every other day in Japanese patients with Type 2 Diabetes Mellitus: a pilot trial. J Med Invest. 2017;64:50-57.
- Nishi A, Numata S, Tajima A, Zhu X, Ito K, Saito A, Kato Y, Kinoshita M, Shimodera S, Ono S, Ochi S, Imamura A, Kurotaki N, Ueno SI, Iwata N, Fukui K, Imoto I, Kamiya A, Ohmori T. De novo non-synonymous TBL1XR1 mutation alters Wnt signaling activity. Sci Rep. 2017;7(1):2887. doi: 10.1038/s41598-017-02792-z.
- Yoshimaru T, Ono M, Bando Y, Chen YA, Mizuguchi K, Komatsu M, Shima H, Imoto I, Izumi K, Honda J, Miyoshi Y, Sasa M, Katagiri T. A-kinase anchoring protein BIG3 coordinates oestrogen signaling in breast cancer cells. Nat Commun. 2017;8:15427. doi: 10.1038/ncomms15427
- Kohmoto T, Okamoto N, Naruto T, Murata C, Ouchi Y, Fujita N, Inagaki H, Satomura S, Okamoto N, Saito M, Masuda K, Kurahashi H, Imoto I*. A case with concurrent duplication, triplication, and uniparental isodisomy at 1q42.12-qter supporting microhomology-mediated break-induced replication model for replicative rearrangements. Mol Cytogenet. 2017;10:15. doi: 10.1186/s13039-017-0316-6.
- Sato Y, Tajima A, Katsurayama M, Nozawa S, Yoshiike M, Koh E, Kanaya J, Namiki M, Matsumiya K, Tsujimura A, Komatsu K, Itoh N, Eguchi J, Imoto I, Yamauchi A, Iwamoto T. An independent validation study of three single nucleotide polymorphisms at the sex hormone-binding globulin locus for testosterone levels identified by genome-wide association studies. Hum Reprod Open. 2017:hox002. Doi. 10.193/hropen/hox002
- Matsudate Y, Naruto T, Hayashi Y, Minami M, Tohyama M, Yokota K, Yamada D, Imoto I, Kubo Y. Targeted exome sequencing and chromosomal microarray for the molecular diagnosis of nevoid basal cell carcinoma syndrome. J Dermatol Sci. 2017;86(3):206-11. doi: 10.1016/j.jdermsci.2017.02.282.
- Shoda K, Ichikawa D, Fujita Y, Masuda K, Hiramoto H, Hamada J, Arita T, Konishi H, Kosuga T, Komatsu S, Shinozaki A, Okamoto K, Imoto I*, Otsuji E. Clinical utility of circulating cell-free Epstein-Barr virus DNA in patients with gastric cancer. Oncotarget. 2017;8(17):28796-28804. doi: 10.18632/oncotarget.15675.
- Kinoshita M, Numata S, Tajima A, Yamamori H, Yasuda Y, Fujimoto M, Watanabe S, Umehara H, Shimodera S, Nakazawa T, Kikuchi M, Nakaya A, Hashimoto H, Imoto I, Hashimoto R, Ohmori T. Effect of Clozapine on DNA Methylation in Peripheral Leukocytes from Patients with Treatment-Resistant Schizophrenia. Int J Mol Sci. 2017;18(3). pii: E632. doi: 10.3390/ijms18030632.
- Kohmoto T, Naruto T, Watanabe M, Fujita Y, Ujiro S, Okamoto N, Horikawa H, Masuda K, Imoto I*. A 590 kb deletion caused by non-allelic homologous recombination between two LINE-1 elements in a patient with mesomelia-synostosis syndrome. Am J Med Genet A. 2017;173(4):1082-1086. doi: 10.1002/ajmg.a.38122.
- Shoda K, Ichikawa D, Fujita Y, Masuda K, Hiramoto H, Hamada J, Arita T, Konishi H, Komatsu S, Shinozaki A, Kakihara N, Okamoto K, Taniguchi H, Imoto I*, Otsuji E. Monitoring the HER2 copy number status in circulating tumor DNA by droplet digital PCR in patients with gastric cancer. Gastric Cancer. 2017;20:126-135. doi. 10.1007/s10120-016-0599-z.
- Kajiura K, Masuda K, Naruto T, Kohmoto T, Watanabe M, Tsuboi M, Takizawa H, Kondo K, Tangoku A, Imoto I*. Frequent silencing of the candidate tumor suppressor TRIM58 by promoter methylation in early-stage lung adenocarcinoma. Oncotarget. 2017;8(2):2890-2905. doi: 10.18632/oncotarget.13761.
- Okamoto N, Watanabe M, Naruto T, Matsuda K, Kohmoto T, Saito M, Masuda K, Imoto I*. Genome-first approach diagnosed Cabezas syndrome via novel CUL4B mutation detection. Hum Genome Var. 2017;4:16045 doi: 10.1038/hgv.2016.45
Education & Training
We welcome you to join our division as graduate school students, research trainees, or senior researchers if you are enthusiastic about cancer research and have background training in fields such as bioinformatics, molecular biology/biochemistry/cell biology, and molecular epidemiology.
Our laboratory is a part of the Nagoya University Graduate School of Medicine. Foreign graduate students are welcome to study in our programs, including those who are supported by the Japanese Government Monbusho (MEXT) scholarship. For details regarding entrance examinations and scholarships, please follow this link:http://admissions.g30.nagoya-u.ac.jp/graduate/.
Recruitment Announcement
Please feel free to contact us.

